Structure and classification of Bacteria
Please click the image for large view in new window
Bacterial classification is important, revealing the identity of an organism so that its behaviour and likely response to treatment can be predicted.
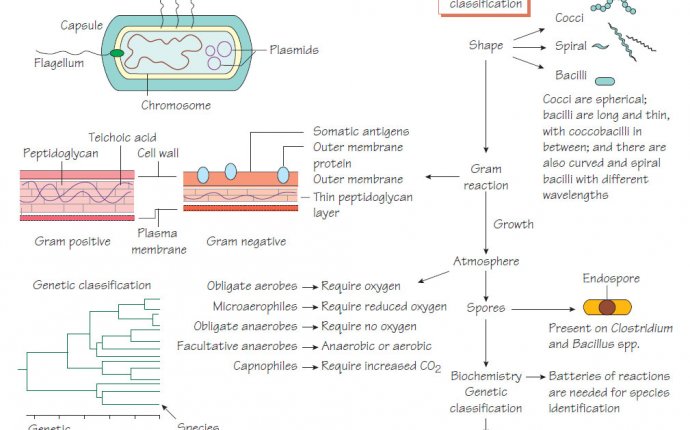
Bacterial structural components
Bacterial cell walls are rigid and protect the organism from differences in osmotic tension between the cell and the environment. Gram-positive cell walls have a thick peptidoglycan layer and a cell membrane, whereas Gram-negative cell walls have three layers: inner and outer membranes, and a thinner peptidoglycan layer. The mycobacterial cell wall has a high proportion of lipid, including immunoreactive antigens. Bacterial cell shape can also be used in classification. The following cell components are important for classification, pathogenicity and therapy.
- Capsule: a polysaccharide layer that protects the cell from phagocytosis and desiccation.
- Lipopolysaccharide: surface antigens that strongly stimulate inflammation and protect Gram-negative bacteria from complement- mediated lysis.
- Fimbriae or pili: specialized thin projections that aid adhesion to host cells. Escherichia coli that cause urinary tract infections bind to mannose receptors on ureteric epithelial cells by their P fimbriae. Fimbrial antigens are often immunogenic but vary between strains so that repeated infections may occur (e.g. Neisseria gonorrhoeae).
- Flagella: these allow organisms to find sources of nutrition and penetrate host mucus. The number and position of flagella may help identification.
- Slime: a polysaccharide material secreted by some bacteria that protects the organism against immune attack and eradication by antibiotics when it is growing in a biofilm in a patient with bronchiectasis or on an inserted medical device.
- Spores: metabolically inert bacterial forms adapted for longterm survival in the environment, which are able to regrow under suitable conditions.
Classification of bacteria
We identify microorganisms to predict their pathogenicity: a Staphylococcus aureus isolated from blood is more likely to be causing disease than Staphylococcus epidermidis. It is important to identify organisms that spread widely in the community and cause serious disease, such as Neisseria meningitidis, so that preventative measures can be taken (see Neisseria and Moraxella ). Bacteria are identified using phenotypic, immunological or molecular characteristics.
- Gram reaction: Gram-positive and Gram-negative bacteria respond to different antibiotics. Other bacteria (e.g. mycobacteria) may require special staining techniques.
- Cell shape: Bacteria may be shaped as cocci, bacilli or spirals.
- Endospore: The presence, shape and position of the endospore within the bacterial cell are noted.
- Fastidiousness: Certain bacteria have specific O2/CO2 requirements, need special media or grow only intracellularly.
- Key enzymes: Some bacteria lack certain enzymes, for example, lack of lactose fermentation helps distinguish salmonellae from E. coli.
- Serological reactions: Interaction of antibodies with surface structures may for example help to distinguish subtypes of salmonellae, Haemophilus and meningococcus.
- DNA sequences: DNA sequencing of key genes (e.g. 16S ribosomal RNA or DNA gyrase) can identify the organism precisely.
Medically important groups of bacteria Gram-positive cocci
are divided into two main groups: the staphylococci (catalase-positive), the major pathogen being S. aureus; and the streptococci (catalase-negative), the major pathogens being Streptococcus pyogenes, which causes sore throat and rheumatic fever, and S. agalactiae, which causes neonatal meningitis and pneumonia (see Streptococcal infections and Streptococcus pneumoniae, other Gram-positive cocci and the alpha-haemolytic streptococci).
Gram-negative cocci include the pathogenic N. meningitidis, an important cause of meningitis and septicaemia, and N. gonorrhoeae, the agent of urethritis (gonorrhoea).