Microbe Classification
 The Baltimore system of virus classification devised by virologist & Nobel laureate David Baltimore is based on the genomic nature of the viruses. The central theme of Baltimore system of virus classification is ‘all viruses must synthesize positive strand mRNAs from their genomes, in order to produce proteins and replicate themselves’. The precise mechanisms whereby this is achieved differ for each virus family.
The Baltimore system of virus classification devised by virologist & Nobel laureate David Baltimore is based on the genomic nature of the viruses. The central theme of Baltimore system of virus classification is ‘all viruses must synthesize positive strand mRNAs from their genomes, in order to produce proteins and replicate themselves’. The precise mechanisms whereby this is achieved differ for each virus family.
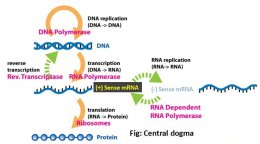
Why + mRNA is placed at the center?
Baltimore classification system is based on the central role of translational machinery and placed mRNA in the center, and described the pathways to form mRNA from DNA or RNA genomes. Viruses can replicate DNA and/or RNA, synthesize RNA from DNA or vice versa, but lack a complete system to make proteins, for which they have to rely on host cell ribosomes. Host cells on the other hand can synthesize proteins only from +mRNA strands. Irrespective of the genomic nature of viruses, all viruses must synthesize viral + mRNAs to produce viral proteins”no exception to date“.
How to designate positive (+) and/or negative (−) strands?
The mRNA which can serve as the template for protein synthesis is defined as a positive (+) strand {by convention (nothing to do with charge/electricity)}. A strand of DNA of the equivalent polarity is also called the (+) strand. RNA and DNA strands that are complementary to the (+) strand are called negative (−) strands.
Viral genomes can be broken down into seven fundamentally different groups, which obviously require different basic strategies for their replication depending on the nature of their genome. When originally conceived, the Baltimore scheme encompassed six classes of viral genome. Subsequently the gapped DNA genome of hepadnaviruses (e.g. hepatitis B virus) was discovered and incorporated in a new scheme.
Viruses can be classified into seven (arbitrary) groups:
I. Double-stranded DNA viruses
Some replicate in the nucleus e.g adenoviruses using cellular proteins. Poxviruses replicate in the cytoplasm and make their own enzymes for nucleic acid replication. e.g. Adenoviruses; Herpesviruses; Poxviruses, etc
II. Single-stranded (+) sense DNA viruses
Replication occurs in the nucleus, involving the formation of a (-) sense strand, which serves as a template for (+)strand RNA and DNA synthesis. e.g., Parvoviruses
III. Double-stranded RNA viruses
These viruses have segmented genomes. Each genome segment is transcribed separately to produce monocistronic mRNAs. e.g., Reoviruses
IV. Single-stranded (+)sense RNA viruses (Picornaviruses; Togaviruses, etc)
- Polycistronic mRNA: Genome RNA = mRNA. Since the RNA is the same sense as mRNA, the RNA alone is infectious, no virion particle associated polymerase. Translation results in the formation of a polyprotein product, which is subsequently cleaved to form the mature proteins. e.g. Picornaviruses (poliovirus, rhinovirus); Hepatitis A virus
- Complex Transcription: Two or more rounds of translation are necessary to produce the genomic RNA. e.g. Picornaviruses; Hepatitis A.
V. Single-stranded (-) sense RNA viruses
The virion RNA is negative sense (complementary to mRNA) and must therefore be copied into the complementary plus-sense mRNA to make proteins. This group of viruses must code for RNA-dependent RNA-polymerase and also carry it in the virion so that they can make mRNAs upon infecting the cell. e.g. Orthomyxoviruses, Rhabdoviruses, etc
- Segmented e.g. Orthomyxoviruses. First step in replication is transcription of the (-) sense RNA genome by the virion RNA-dependent RNA polymerase to produce monocistronic mRNAs, which also serve as the template for genome replication.
- Non-segmented e.g. Rhabdoviruses. Replication occurs as above and monocistronic mRNAs are produced.
VI. Single-stranded (+) sense RNA viruses with DNA intermediate in life-cycle
RNA genome is (+) sense but unique among viruses in that it is DIPLOID, and does not serve as mRNA, but as a template for reverse transcription. e.g. Retroviruses.
Retroviruses therefore encodes an RNA-dependent DNA polymerase (reverse transcriptase) to make the DNA provirus which then is transcribed to genomic RNA by a host enzyme, RNA polymerase II.
VII. Double-stranded DNA viruses with RNA intermediate
This group of viruses also relies on reverse transcription, but unlike the Retroviruses, this occurs inside the virus particle on maturation. On infection of a new cell, the first event to occur is repair of the gapped genome, followed by transcription. e.g., Hepadnaviruses